insights
The Functional Medicine Approach to COVID-19: Primer on SARS-CoV-2 Testing
Published: 04/23/20
Introduction
The ability to accurately identify whether individuals are at risk for, infected with, or have an immune response to SARS-CoV-2 is essential to address the COVID-19 pandemic from both a personal and a public health perspective. Reliable testing is critical for:
- Determining the state of an individual’s immune response to exposure.
- Assessing whether an individual is contagious and needs to be quarantined.
- Deciding if an individual is immune and can return to society without undue risk.
- Facilitating contact tracing to reduce viral spread.
- Determining the prevalence and natural history of this novel disease.
The purpose of this document is to help clinicians understand the important issues regarding testing, including:
- The types of testing available.
- Assessment of the reliability of these tests.
- Using test results to provide a framework for clinical decision–making for patients.
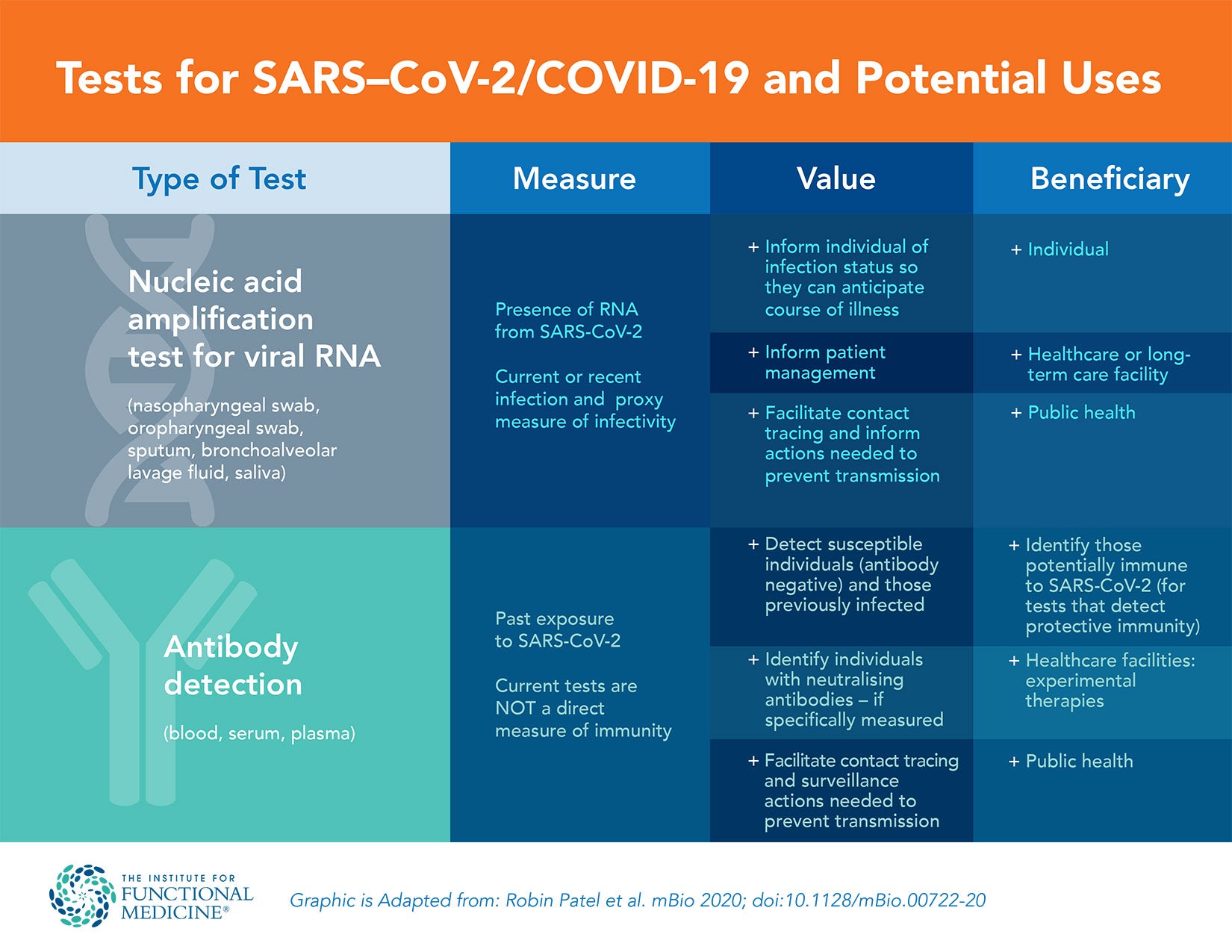
Laboratory testing for SARS-CoV-2 includes two main categories: those that detect the presence of the virus and those that determine the host response to the virus by detecting antibodies specific to SARS-CoV-2.2 Transmission of SARS-CoV-2 can occur from direct contact or via airborne droplets. There are several possible responses after an individual is exposed to the virus, as shown in the table below. A similar table is included in the summary, with inclusion of clinical interpretation and recommended action.
| Possible Outcomes After Exposure to SARS-CoV-2 | ||
|---|---|---|
| Symptomatic | Viral RNA shedding | Production of antibodies |
- |
- |
- |
- |
+ |
- |
- |
- |
+ |
- |
+ |
+ |
+ |
- |
- |
+ |
+ |
- |
+ |
- |
+ |
+ |
+ |
+ |
Time Course
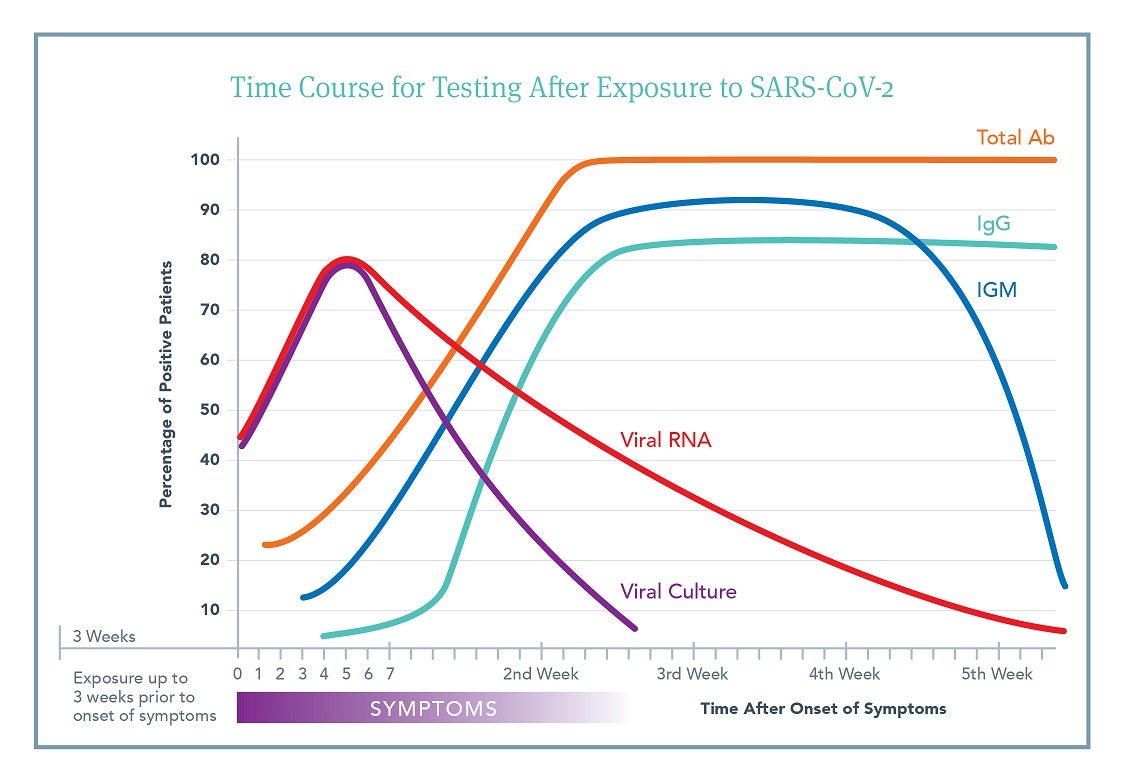
Although there is a vast amount of individual variability in response to viral exposure, understanding the general time course of exposure, onset of symptoms, length of viral shedding, and production of antibodies is important for deciding which test to order and how to interpret the subsequent results.
This figure is an aggregate view based on a number of different published studies.3,4,5,6,7,8,9,10 It outlines the likelihood that an individual patient will have a positive test result at a specific time after onset of symptoms. Day 0 is time of symptom onset. Click to Download PDF
Time of exposure to symptom development ranges from 2-21 days.
- >90% of people who develop symptoms will do so within 14 days.
- Up to 10% of people may develop symptoms later than 14 days.
- Many people never develop symptoms but may still develop antibody responses. The timing and level of antibody responses in asymptomatic people is presently unknown.
Viral shedding can begin 3-21 days after exposure (i.e., preceding symptoms).
- Viral RNA shedding can occur in nasopharynx, oropharynx, saliva, and sputum for up to 21 days after symptom resolution.
- Testing evaluates viral RNA, not shedding of intact virus (i.e., infectivity).
- It appears that intact viral shedding is complete 14 days after symptoms begin, though viral RNA shedding continues (as noted above).
Antibody production begins soon after symptoms (5-14 days).
- Timing of antibody development (IgM, IgG, & IgA) varies across individuals.
- Median antibody production is at 15 days after exposure.
Accuracy
Before assessing the specific tests in more detail, it is important to understand the accuracy of a test; in other words, how likely is a test to return a true result? We measure the accuracy of a diagnostic test via the following characteristics:
- First, sensitivity, which is the percentage of truly infected individuals who will test positive on a test.
- Second, specificity, which is the percentage of truly uninfected individuals who test negative.
The sensitivity of a test shows how well a test detects the proportion of true positives (TP); the probability that the test will capture a positive result and avoid a false negative (FN) result: (TP/TP+FN). Clinically, using serology tests with high sensitivity ensures that clinicians do not falsely miss the presence of antibodies to the virus.
The specificity of a test shows how well a test detects the proportion of true negatives (TN); the probability that the test will not give a false positive (FP) result: (TN/TN+FP). Clinically, using RNA nucleic acid amplification (NAA) tests with high specificity ensures that clinicians do not falsely assume the presence of the virus.
Additional characteristics of diagnostic tests that are important to consider are positive predictive value, or PPV, and negative predictive value, or NPV. Positive predictive value is defined as the percentage of those testing positive on a test who are truly infected. Put another way, if you have someone who tests positive, the positive predictive value is the percent chance that that person actually does have the infection. Negative predictive value is defined as the percentage of those testing negative on a test who are truly uninfected. Both PPV and NPV depend not only on the sensitivity and specificity of the test but also on the prevalence of infection in the population studied.
Issues of sampling error with antigen testing (insufficient or incorrect sample procurement) are noted below and are independent of the sensitivity and specificity of the assay.
Understanding Testing
Viral Testing
Viral RNA
RT-PCR Techniques
Tests that look for the presence of the SARS-CoV-2 in the respiratory tract are used to determine if an individual has an active infection and/or is contagious. Most of the tests currently available for this purpose detect the presence of viral RNA. SARS-CoV-2 has an RNA genome; thus, determining if RNA sequences specific to the known SARS-CoV-2 genome are present can determine if viral particles are present in the sample. This test is not a direct measure of whole virus, which would have to be evaluated with viral culture. Viral RNA detection is most often achieved via nucleic acid amplification methods (NAA) using a polymerase chain reaction (PCR). Samples are collected from nasopharyngeal (NP) swab, oropharyngeal (OP) swab, bronchoalveolar lavage, sputum (research), or saliva and then placed in medium that preserves the RNA.
At the lab, RNA is isolated from the sample and then subjected to a reverse transcription reaction to convert the RNA sequence to DNA. Specific part(s) of the sequence are then amplified using real-time reverse transcription PCR (RT-PCR). The RT-PCR technique uses primers, which are short fragments of DNA that are complementary to specific parts of the transcribed viral RNA. These primers are added to the reaction with appropriate enzymes that amplify the target sequence hundreds to thousands of times through successive cycles of heating and cooling, making it easier to identify whether the specific sequence is present in a large amount of other genetic information such as host DNA. The RT-PCR technique uses fluorescently labeled DNA strands to monitor the amplification reaction and identify the presence of the specific strand in “real-time” as the reaction proceeds. It has the advantage over conventional PCR in that it takes place in a single reaction, resulting in less chance for error and a shorter completion time. It has the additional advantage of being quantitative, thus giving an estimate of viral load (i.e., quantity). Simultaneously performing multiplex RT-PCR, which tests for multiple target sequences, increases both sensitivity and specificity.11
Individual laboratory assays have different cut-offs for the amount of RNA that defines a positive result. This will affect the sensitivity and specificity of the assay. Thus, test performance characteristics and cut-offs are laboratory-dependent and will affect clinical and epidemiological interpretation. Finally, despite the ability to quantify the viral RNA in the initial sample, most results are reported as a qualitative presence or absence of viral RNA. Quantitative results are primarily used in the research setting at this time.
CRISPR-Cas12
In addition to the widely adopted RT-PCR discussed above, new methods also include the use of CRISPR-Cas12 gene targeting technology to test for the presence of SARS-CoV-2 RNA in OP & NP swabs. CRISPR-Cas12 can be modified to target any genetic sequence—for example, targeting two regions in the genome, one that is unique to SARS-CoV-2 and one that is common to all SARS-like coronaviruses. This ensures the assay is distinguishing between SARS-CoV-2 and closely related viruses. The test also decreases turnaround time by cutting a four-hour-long assay down to 45 minutes, thus improving the speed of diagnosis.12
Point-of-Care Tests
Recently, a number of companies have been developing rapid point-of-care (POC) tests that can detect viral nucleic acids or viral proteins (antigens). Rapid POC tests that will make individual testing more readily available are eagerly anticipated. The results of proper validation studies for these tests are essential before their widespread adoption.
A handheld DNA analyzer that automates nucleic acid extraction and amplification has recently been repurposed for SARS-CoV-2 detection. It provides results in approximately one hour and can be used at point-of-care locations.13 It has been approved by Health Canada but is not yet widely available.
Lateral flow immunoassays that can detect various SARS-CoV-2 proteins have also been developed. The lateral flow assay is similar to a home pregnancy kit in which “capture reagents,” such as monoclonal antibodies (mAbs), are directed at a viral antigen and are immobilized at defined locations on a nitrocellulose membrane along with labeled detector mAbs to the same target. Binding between the analyte (viral protein or RNA in sample) and the capture and detector mAbs reveals a positive test by producing a visible colored line.11
Viral Culture
Assessing the ability of the virus collected from a sample to grow in culture can determine whether the sample contains infectious viral particles as opposed to only viral RNA. These assays are time consuming and require labs with high-level biohazard containment facilities and are best reserved for research purposes.
Sample Collection
The accuracy of results using viral detection tests are most often contingent on the type of sample being tested and the methodology of collection. Viral RNA can be extracted from all types of respiratory membranes, including OP, nasal, and NP swabs; saliva; and sputum. One smaller study found no difference in detection rates between OP and NP,14 while another larger study found that sputum was most accurate, followed by nasal swabs for detecting SARS-CoV-2 RNA. While sputum, nasal, and throat swabs revealed positive results, after eight days, the positive rate for throat swabs was much lower.15 A more recent study showed that saliva from the posterior oropharynx (coughed up by clearing the throat), especially first thing in the morning after being supine all night, also produced high yield samples that mirrored endotracheal aspirates. Notably, the non-invasive nature of saliva collection may be more acceptable to patients requiring serial sampling.7 Currently, best evidence suggests that when given a choice, sputum followed by nasal-pharyngeal (NP) swab and saliva (FDA approved on April 18, 2020) are the preferred sample types. In addition to the type of sample collected, proper methodology during sample collection is critical.
RT-PCR is estimated to have up to a 30% false negative rate for identifying viral RNA in an infected person. The majority of this is the result of collection procedures that use inappropriate sites or collect inadequate amounts of material. Collection outside the diagnostic window (3-21 days after exposure) can also increase the false negative rate. Handling, transport, and storage of swabs as well as the presence of interfering substances or contamination also play a role in the false negative rate.16
One study showed that the titers of viral RNA tend to peak at or just before symptom onset followed by a rapid decrease in the naso- and oropharynx and a gradual decrease in sputum and stool. Titers remain high, especially in the sputum and stool, even after symptoms resolve.3 This begs the question as to whether the presence of viral RNA in a sample equates to infectious viral shedding. The answer appears to be “No,” since it was shown that even though stool samples can contain high amounts of viral RNA, infectious viral particles grown in culture have not been isolated from stool.3 Even though the presence of viral RNA is not perfectly correlated with infectious viral shedding, it is prudent to assume that a patient with a positive respiratory RT-PCR test is contagious and should observe appropriate quarantine measures.
Regulations
Labs performing viral RNA testing must have emergency use authorization (EUA) from the FDA and should have CLIA certification for high complexity testing. Since there are a large number of labs offering testing, it is important to ensure they are meeting appropriate regulatory guidelines. Currently, all POC testing must be done in a CLIA-certified lab (https://www.cdc.gov/coronavirus/2019-nCoV/hcp/clinical-criteria.html).
| Viral Testing Summary | |
|---|---|
| Samples | Sputum, if available, then nasopharyngeal, nasal, and lastly, oropharyngeal swabs. Saliva testing is now also available and works best when collected from a deep cough after arising. Take samples within seven days of symptom onset. |
| Methods | Ensure viral transport medium is not expired and follow proper collection techniques. Send for RT-PCR at FDA, EUA-approved CLIA lab. |
| Timing | Viral shedding can begin 3-21 days after exposure. Viral RNA shedding can occur for up to 21 days after symptom resolution. Testing evaluates viral RNA, not shedding of intact virus (i.e., infectivity). It appears that intact viral shedding is, in most cases, complete 14 days after symptom onset—note that this differs from the time course of viral RNA shedding, as measured by NAA. |
Antibody Testing
Serological Testing
Serological testing is used for assessing an individual’s immune response to SARS-CoV-2 exposure. The addition of serological testing to viral RNA testing dramatically increases the sensitivity of a COVID-19 diagnosis and facilitates assessment of past exposure (especially in asymptomatic cases).4,8,17 Serological testing from blood, serum, or plasma determines if one or more immunoglobulin subtypes (IgM, IgA, or IgG) are present to specific viral antigens.
The development of specific antibodies to different protein components of a virus occurs in a sequential manner, with IgM and IgA arising initially, followed by IgG, which confers long-term immunity.18 IgM tends to be the more general, immediate response immunoglobulin, while subsequent isotype switching to IgG selects for antibodies with higher binding affinity. IgA is most often found at the mucous membranes. Current evidence supports this immunological paradigm in SARS-CoV-2 infection as well.3,5,10 While it appears that close to 100% of individuals who are infected (PCR positive) go on to develop some type of antibody response to the virus, there is tremendous individual variability in the time course of antibody production, the amount of antibody produced, and the specificity of the antibodies, i.e., which viral proteins are recognized. Antibody levels also appear to correlate with severity of disease and presence of comorbidities. More severe disease results in higher viral load and also higher antibody titers.5 An individual’s underlying immune competence, such as specific immunoglobulin deficiencies, will also affect whether they are capable of mounting an efficient immune response resulting in positive antibody titers.
As immunoassays are developed, it becomes important to determine the most appropriate viral antigens to use. There are many companies currently developing serology testing, and many are not forthcoming as to the antigen being used in their assay. Recent studies have looked at the whole spike protein (S) or the S1, S2, or receptor-binding domain (RBD) subunits of S, in addition to the nucleoprotein (N) and the membrane protein (M). The S and N proteins are the main immunogenic proteins, while S1 and RBD correlate with neutralizing antibodies (NAbs), since they are the proteins on the viral surface that facilitate entry into the host cell.6,7 Neutralizing antibodies not only bind to a viral protein but are capable of blocking a viral infection by neutralizing or inhibiting its biological effect. This is opposed to binding Abs, which bind to a specific antigen flagging them. The antibody-antigen complex can then trigger T cells to destroy the flagged antigen.6 NAbs can directly inhibit viral infection without the need for further cellular immune support, and are thus key to preventing subsequent infection. This explains why therapeutic convalescent plasma is effective if high levels of NAbs are present.
The choice of viral antigen used for the immunoassay is also important because other coronaviruses have similar proteins, and an immune response to a previous coronavirus may cross-react, resulting in a false positive test (specificity). Not all individuals will mount an Ab response to every viral antigen, making the choice of viral antigen also important for increasing the sensitivity of the test. Thus, the level of sensitivity and specificity of each test will vary with the antigen used, the serotype of the antibody being tested, and the parameters of the assay itself.1
Despite immense variability, most individuals develop some type of seropositivity within seven to ten days following onset of symptoms. Testing prior to seven days post symptom onset is likely to yield a negative antibody result. In one study, the positive rate for IgG or IgM at less than seven days was 38%, while the rate at greater than seven days was 89%.3 Another study in 23 hospitalized patients revealed that by day 14, rates of seropositivity were 94% for anti-NP IgG, 88% for anti-NP IgM, 100% for anti-RBD IgG, and 94% for anti-RBD IgM,7 indicating that testing later in the time course of the disease is more sensitive and that there is variability in the immunoglobulin isotype and the viral antigens.
Types of Antibody Testing
Laboratory Standard = ELISA Testing
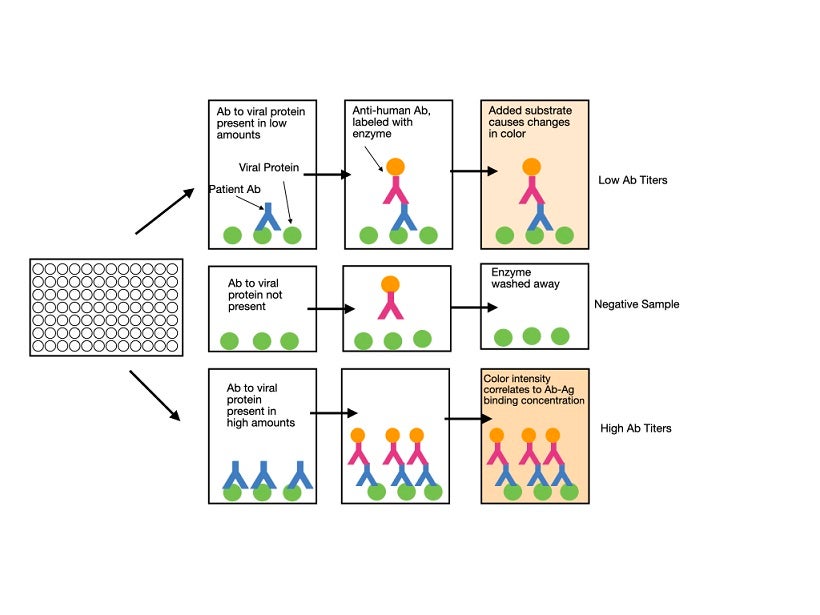
Enzyme-linked immunosorbent assay (ELISA) is a plate-based technique that uses whole blood, plasma, or serum samples from patients for testing. The antigen of interest (for example, the S protein) is coated on the surface of the plate. The patient’s sample is then added. If the sample contains antibodies that recognize the antigen, they will bind to it, while other antibodies are washed away. A labeled (usually fluorescent) detection antibody that recognizes a specific immunoglobulin isotype is subsequently added. Binding of the antibody in a positive sample is detected by measuring the florescent-based readout. The higher the concentration of specific antibody in the sample, the stronger the signal will be. Although most labs will purchase pre-coated plates, they should still perform their own validation studies to ensure that the test produces accurate results. An ELISA typically takes one to five hours to perform and can determine the quantitative presence or absence of antibodies against the virus in the patient’s blood. It does not, at this point in time, give information as to whether the antibodies are able to protect against future infection.
Rapid Diagnostic Testing
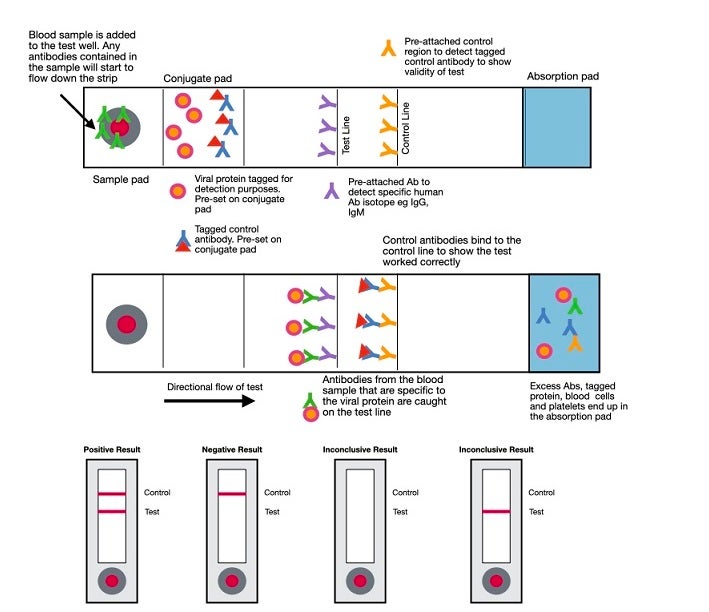
Rapid diagnostic tests (RDT) are typically created to yield a qualitative result of presence or absence. While most are currently being performed in labs, they lend themselves well to point-of-care testing. RDTs for antibodies are lateral flow assays and use the same principles as the lateral flow assays used for antigen detection discussed above. In this case, a labeled antigen such as the S (spike) protein is at the bottom of the cassette. The patient sample is added to the membrane, and if antibodies to the labeled protein are present, they will bind to the protein and be captured by an immobilized isotype-specific antibody, resulting in a visible line at the capture site. As of this date (April 22, 2020), point-of-care tests are only allowed to be performed in CLIA-certified laboratories that have emergency use authorization (EUA) to run the test. Clinicians performing POC tests in their office are operating outside of regulatory guidelines.
Click to Download PDF
Validation
Laboratory companies are moving quickly to expand the ability to test. While this is much needed, it can also lead to a lack of sufficient validation to ensure trust in the result. Approval under EUA, while important, does not provide the level of rigor required for an FDA-approved test. In this environment, it falls on the clinician to make their own determination of the value of the test(s) they choose to use.
Each lab that offers testing should be forthcoming with their internal process of analytical validation. CLIA certification is important and ensures that a test meets analytical validity. Analytical validity ensures the correct response for the sample tested. High reproducibility and low coefficient of variation fall into this category (i.e., when the same sample is tested repeatedly, it should produce the same result).
Clinical validity, on the other hand, is the correct response for the patient tested rather than the sample.19 In order to ensure clinical validity, one must ask a number of questions about the validation process, including:
- Where are you obtaining the kits or reagents to do your testing? (Most RDT kits are currently purchased from China).
- What are the control samples? Is the lab using clinical samples as opposed to ‘spiked’ (i.e., artificial) samples?
- Is the lab using confirmed SARS-CoV-2 positive samples, and if so, how and when was this confirmed?
- When was the sample collected for antibody testing in relation to a positive viral RNA test?
- What are the demographics of the clinical test population?
- Did the positive test population have symptoms in addition to a positive PCR test?
- Were pre-pandemic samples used in the negative test group?
- Did the assay evaluate samples that had exposure to other respiratory viruses such as influenza and RSV, as well as other coronaviruses, to determine cross reactivity and specificity?
- How many samples were used in the validation?
- Is the lab doing progressive validation studies after the test is released?
As the development of serological testing moves forward, it will be critical to use well-defined standard references as part of diagnostic assay validation in order to standardize assays developed by different labs to better understand correlates of immune protection.10 In the interim, the choice of the most appropriate test falls to the individual labs and clinicians using those labs to ensure validity.
There are still a number of unknowns, such as:
- Are individuals infected with SARS-CoV-2 who recover protected either fully or partially from future infection?2
- Do all immunocompetent individuals mount an antibody response to all the viral proteins?
- How long do immunocompromised individuals experience intact viral shedding?
- Will future viral mutations change the sensitivity and specificity of testing and the development of immunity?
- Does having antibodies prevent community spread?
- Does having antibodies confer immunity and prevent recurrence?
- How long might protective immunity last?
A study done in rhesus macaques sheds some light on the question of immunity. These animals show similar patterns of infection and COVID symptoms from SARS-CoV-2 as humans. Viral replication in the nose, pharynx, lung, and gut, as well as interstitial pneumonia, were seen after primary infection. Once symptoms resolved and they demonstrated a positive Ab test, the monkeys were re-challenged with the same dose of SARS-CoV-2. No signs of infection, including viral replication in any of the tissues, were seen in the re-exposed animals.20 This gives some basis for the assumption that infection followed by positive antibody titers very likely confers some type of immunity, although this has yet to be proven in humans.
Regulations
The current COVID-19 pandemic is a public health emergency and justifies the declaration of emergency use authorization (EUA) of in vitro diagnostics for the detection and/or diagnosis of SARS-CoV-2. Labs seeking EUA for their test must be CLIA certified in order to make a submission to the FDA. After expedited review, the FDA can grant EUA for a test when there is reasonable belief that the test may be effective in diagnosing COVID-19, that the known or potential benefits of using the test outweigh the potential risks, and that there are no adequate, approved, alternatives.
BUYER BEWARE: The FDA outlines a number of conditions when it grants EUA, including requiring the lab to clearly state that the test has not been FDA cleared or approved, that it is authorized by FDA under an EUA, and that it is only authorized for use for the duration of the declaration that circumstances exist justifying emergency use. Full FDA approval has not been granted to any lab, test, or product for SARS-CoV-2. Labs claiming that they have “FDA approval” are doing so in contradiction of their EUA status.
As per the FDA website on April 20, 2020, labs that perform antibody testing are temporarily not required to possess an FDA EUA. The FDA encourages, but does not require, labs that offer this testing to submit an EUA application. The FDA states that validation studies are important in judging the quality of a test and will be required for an EUA. Therefore, we recommend that, ideally, clinicians use antibody tests that have an FDA EUA but, in the absence of such, ensure that quality validation studies were done.
The astute clinician understands that while there is a certain amount of regulatory oversight with EUA, it does not constitute formal federal review and approval of the test or of the validation process. Manufacturers of tests and the labs that utilize them are being given a level of trust that is needed and hopefully justified.
| Summary of Antibody Testing | |
|---|---|
| Samples | Whole blood, plasma, or serum. |
| Methods | Send to lab with EUA and validated ELISA or lateral flow membrane testing. Point-of-care testing is not currently FDA-approved (as of April 20, 2020). |
| Timing | Higher sensitivity begins seven days after symptom onset. |
| Type | Testing for both IgM and IgG antibodies measuring multiple viral proteins will increase both sensitivity and specificity. |
Interpretation
If the clinician has a high index of suspicion that a patient is likely to be or has been infected with SARS-CoV-2, and testing returns a negative result, the following questions may help to determine whether the test should be repeated.
If negative NAA:
- Was the swab collected correctly?
- Was a sputum, saliva, or NP sample collected?
- Is the patient symptomatic?
- Does the patient have a known exposure to a person with a positive test?
- Where is the patient in the time course of the illness (i.e., are they beyond the first week, when viral RNA levels decline)?
If negative antibody:
- Does the patient have immunoglobulin deficiency or are they immunocompromised?
- Did the patient have a positive NAA viral test?
- Was the sample collected greater than 7-14 days after symptom onset?
- What isotype is being tested: IgM or IgG? (If longer time since exposure, IgM is less likely positive, and if more recent, IgG is less likely positive).
- What was the sensitivity of the test being used?
| Symptomatic | Viral RNA shedding | Production of antibodies | Interpretation | Action |
|---|---|---|---|---|
- |
- |
- |
Susceptible | Social isolation to prevent contracting virus; risk stratification to determine return to work (based upon public policy). |
- |
+ |
- |
Infected | Quarantine, retest RNA and Ab in two weeks. |
- |
- |
+ |
Past exposure, immunity likely, evaluate for return to work | If an individual had symptoms, then RNA and Ab test seven days after resolution of symptoms. Return to work if RNA negative and Ab positive. If individual never had symptoms, repeat RNA test and Ab test and consider return to work if RNA negative and Ab positive. |
- |
+ |
+ |
Infected with developing immunity | Quarantine, retest RNA and Ab in seven days, return to work if RNA negative and Ab positive (consider additional RNA negative test if exposure to high risk populations). |
+ |
- |
- |
Assume infected | Quarantine, consider second RNA test to confirm. |
+ |
+ |
- |
Infected | Quarantine, retest RNA and Ab in two weeks. |
+ |
- |
+ |
Infected with developing immunity | Quarantine, consider second RNA test to confirm. |
+ |
+ |
+ |
Infected, with developing immunity | Quarantine, retest RNA and Ab in two weeks. |
- This table indicates interpretation of possible testing scenarios and suggested actions.
- Note that return to work will be dictated by local public policy.
- The need for repeat confirmatory testing is suggested based on the sensitivity and specificity of currently available tests, which are often not close to 100%.
- These are guidelines only. The clinician must use their judgment, based on their knowledge of the test that is used.
Clinical Scenarios
As the clinician attempts to navigate an individual patient through the muddy waters of the current testing environment, it may help to imagine how one might approach a number of different clinical scenarios with respect to testing.
If a patient had:
- A known exposure but was asymptomatic:
- Quarantine for two weeks, then test for Abs and viral RNA two weeks post-exposure and follow the suggestions in the above table based on the results of the test.
- Mild or non-classic symptoms:
- Perform viral RNA test to confirm infection.
- If viral RNA is negative, then perform Ab test seven days after symptoms resolve, with confirmatory Ab test in one to two weeks.
- If viral RNA is positive, perform Ab test three to seven days after symptoms resolve, with confirmatory Ab test in one to two weeks.
- It is prudent to confirm a second negative RNA test prior to considering return to work.
- Classic symptoms:
- Perform viral RNA test to confirm infection.
- If viral RNA negative, perform Ab test three to seven days after symptoms resolve, with confirmatory Ab test in one to two
- If viral RNA positive, then viral RNA and Ab test seven days after symptoms resolve, with confirmatory Ab test in one to two
- It is prudent to confirm a second negative RNA test prior to considering return to work.
Summary
The understanding of SARS-CoV-2 is continually evolving as more is learned about the virus and the host’s response to exposure and infection. Testing is also evolving as this greater understanding is combined with rapid advances in technology to create tests for both the virus and the immune response.
The clinician on the front lines of this pandemic will find this document helpful to understand which tests are currently available, what circumstances warrant their use, and how to interpret and act on the results. This document will also help clinicians to have an understanding of testing methodologies, validation processes, and the regulatory environment necessary to assess new tests as they become available.
Click here to view and download the laboratory request for information form, Assessment of Accuracy, Analytical and Clinical Validity of Lab Testing.
Webinar
SPECIAL THANKS
We would like to thank the IFM COVID-19 Task Force, members of the IFM staff, and consultants working with IFM for their contributions to this article.
References
- Petherick A. Developing antibody tests for SARS-CoV-2.?Lancet. 2020;395(10230):1101-1102. doi:1016/S0140-6736(20)30788-1
- Patel R, Babady E, Theel ES, et al. Report from the American Society for Microbiology COVID-19 International Summit, 23 March 2020: value of diagnostic testing for SARS-CoV-2/COVID-19.?mBio. 2020;11(2):e00722-20. doi:1128/mBio.00722-20
- Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. Published online April 1, 2020. doi:1038/s41586-020-2196-x
- Lou B, Li T, Zheng S, et al. Serology characteristics of SARS-CoV-2 infection since the exposure and post symptoms onset. medRxiv. Published online March 27, 2020. doi:1101/2020.03.23.20041707
- Zhao J, Yuan Q, Wang H, et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019.?Clin Infect Dis. Published online March 28, 2020. doi:1093/cid/ciaa344
- Wu F, Wang A, Liu M, et.al. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. medRxiv. Published online April 20, 2020. doi:1101/2020.03.30.20047365
- To KK, Tsang OT, Leung WS, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. Published online March 23, 2020. doi:1016/S1473-3099(20)30196-1
- Guo L, Ren L, Yang S, et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19).?Clin Infect Dis. Published online March 21, 2020. doi:1093/cid/ciaa310
- Chang D, Mo G, Yuan X, et al. Time kinetics of viral clearance and resolution of symptoms in novel coronavirus infection.?Am J Respir Crit Care Med. Published online March 23, 2020. doi:1164/rccm.202003-0524LE
- Okba NMA, Müller MA, Li W, et al. Severe acute respiratory syndrome coronavirus 2–specific antibody responses in coronavirus disease 2019 patients.?Emerg Infect Dis. 2020;26(7). doi:3201/eid2607.200841
- Sheridan C. Fast, portable tests come online to curb coronavirus pandemic.?Nat Biotechnol. Published online March 23, 2020. doi:1038/d41587-020-00010-2
- Broughton JP, Deng X, Yu G, et al. CRISPR-Cas12-based detection of SARS-CoV-2.?Nat Biotechnol. Published online April 16, 2020. doi:1038/s41587-020-0513-4
- Spartan COVID-19 test: rapid identification of the COVID-19 virus. Spartan. Published 2020. Accessed April 21, 2020. https://www.spartanbio.com/products/medical/covid-19/
- Woelfel R, Corman VM, Guggemos W, et al. Clinical presentation and virologicalassessment of hospitalized cases of coronavirus disease 2019 in a travel-associated transmission cluster.?Published online March 8, 2020.?doi:10.1101/2020.03.05.20030502
- Yang Y, Yang M, Shen C, et al. Evaluating the accuracy of different respiratory specimens in the laboratory diagnosis and monitoring the viral shedding of 2019-nCoV infections. Published online February 17, 2020. doi:10.1101/2020.02.11.20021493
- Lippi G, Simundic AM, Plebani M. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (COVID-19).?Clin Chem Lab Med. Published online March 16, 2020. doi:1515/cclm-2020-0285
- Zhang W, Du RH, Li B, et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes.?Emerg Microbes Infect. 2020;9(1):386-389. doi:1080/22221751.2020.1729071
- Li ZN, Lin SC, Carney PJ, et al. IgM, IgG, and IgA antibody responses to influenza A(H1N1)pdm09 hemagglutinin in infected persons during the first wave of the 2009 pandemic in the United States.?Clin Vaccine Immunol. 2014;21(8):1054-1060. doi:1128/CVI.00129-14
- Palomaki GE, Haddow JE. Confusion between analytic validity and clinical?validity.?Am J Obstet Gynecol. 2016;215(4):533-534. doi:1016/j.ajog.2016.06.008
- Bao L, Deng W, Gao H, et al. Reinfection could not occur in SARS-CoV-2 infected rhesus macaques. bioRxiv. Published online March 14,